Whole Genome Profiling 2.0
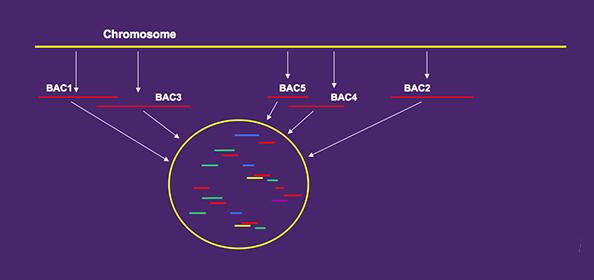
With WGP technology you get high quality sequence tags from BAC’s assembled into contigs.

| Tomato Genome Improvements! | WGP Original Version | WGP 2.0 |
|---|---|---|
| Published in Nature May 31, 2012 |
Chimeric BAC removal & SuperBAC strategy |
|
| # BACs | 66,084 | 54,855 |
| # SuperBACs & remaining single BACs | n/a | 14,683 & 1956 |
| # tags | 261,913 | 255,341 |
| # (Super)BACs in contigs | 52,617 (80%) | 15,175 (91%) |
| # contigs | 251 | 665 |
| N50 contig size (Mb) | 0.563 Mb | 3.312 Mb |
| Avg contig size (Mb) | 0.378 | 1.323 |
| Coverage (Mb) | 953 | 880 |
Experience with various WGP Projects:
The patented WGP technology was published in Genome Research; you can also read our chapter on WGP in Tag-Based Next Generation Sequencing (Chapter 17).
We have performed WGP on a 4.5 Gbp Tobacco strain (details were presented by our client at PAG in 2011), two strains of Arabidopsis, a 450 Mbp melon (de-novo), a 150 Mbp parasite, and dozens of other plant species. Our clients have presented WGP data on the potato and tomato genome (Click here to read more PDF). We have performed WGP on BAC clones from very large genomes and have found WGP to be “scaleable” on huge genomes. The Whole Genome Profiling WGP Results 9 Projects Amplicon and KeyGene PDF includes details on nine different WGP projects from various plants.
Amplicon Express and Keygene have generated over 40 physical maps for 27 different species with varying genome sizes (130 Mbp -5000 Mbp) and complexities. New pop-up window WGP track record.
Why WGP 2.0 is better than SnapShot or other mapping/fingerprinting technologies:
With WGP, BAC clones are assembled into contigs which are linked together by DNA sequence data, not by DNA fragment lengths as with other fingerprinting technologies! Fragment-length data are not sequence based, so SnapShot data cannot be added/assembled directly into a Whole Genome Sequencing project.
Contact us to get a WGP 2.0 quote.